15 Enzyme compartmentalization in cells and tissues
1. Objectives
a. Enzyme compartmentation
b. Various methods for studying compartmentation
c. Metabolic pathways and enzyme compartmentation
2. Description on Enzyme compartmentalization
Enzymes, that catalyze metabolic pathways are very well compartmentalized for its action-either inter or intra cellular compartmentation.This division of role played is assisted or supported by other cellular components like energy supplies(ATP/GTP),various metabolic precursors or intermediates. All the compartments rely on the master organelle ‘nucleus’ and ‘proteinfactories’-the ribosomes to provide most or all its components. Its interguing to understand how the complexity of eukaryotic system is maintained through well compartmentalized process and how they link together. Other than compartmentation,isoforms or isozymes are another class, which alters the catalytic and regulatory aspect of enzymes. They normally present in different tissues, cells, organelles or alter their locations within the cell under different physiological conditions like abiotic stress etc.Thus compartmentation of metabolic tasks organization function and regulation.
Eg: Enormous change in muscle metabolic rate during excess movement in animals, various plant metabolic processes mediated by different organelles like chloroplast, mitochondria and peroxisome.
3. Various methods used for studying compartmentation:
It’s surprising to understand how cell or tissue compartmentalizes its metabolic need. Various traditional and modern methods have been used to unravel and to understand the localization and regulation aspects of cell compartmentation.
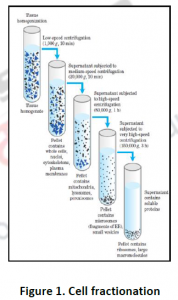
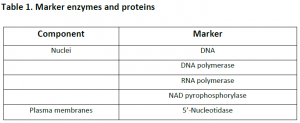
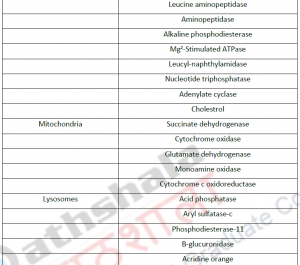
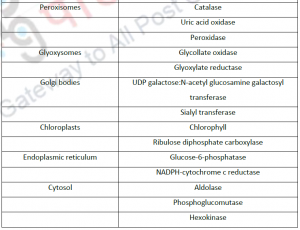
3.1. Cell fractionation: For separating various organelles from cells as well as various cells from tissues, gentle disruption in an osmotically stable medium is mandatory. Cell lysate is further fractionated by differential or density gradient centrifugation using sucrose,caesium chloride,ficoll or percoll gradients (Figure 1). Each fraction can then collected and assayed for particular enzyme activities of marker enzyme for each compartment. Some enzymes may colocalize across different fractions (Table 1). For further experiments on highly purified organelle preparations can also be carried out to confirm the location of enzyme. Latency measurement of enzymes is another most common biochemical assay in which the activity is compared between lysed and intact organelle fractions. Protease protection assays are another commonly performed where resistance to proteolytic degradation untill the organelle is broken,which indicates that the enzyme is located inside.The major limitation of cell fractionation technique is that, the cytosolic location of an enzyme can only be found or referred in a negative way i.e. if it doesn’t colocalize to any known organelle.
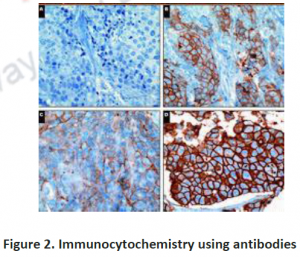
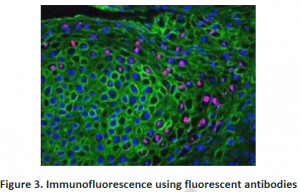
3.2. Immunohistochemistry: To investigate compartmentation, a histology based staining method is commonly performed. The word ‘immuno’ denotes the use of antibodies, which are specific against particular proteins and then visualized by light or electron microscopy after labeling with fluorescent, luminescent, radioactive or enzymatic markers (Figure 2 and 3). This technique is mostly used for those proteins which do not have enzymatic activity especially transporters or unknown proteins etc. Histo-chemistry can also used to locate non-protein targets such as cell-wall polysacharides and amino acids. Immuno- histochemical methods can overcome the limitations of cell fractionation of cytosolic proteins can be distinguished from those that are associated with intracellular structures that are too fragile or too low in abundance like ER,golgi etc. A major limitation of this antibody based technique is the need for identifying specific antibodies for each target of interest.Occasionally weak cross reaction with certain abundant proteins(like Rubisco) can lead to misinterpretations. Antibodies, though they are structure based interactions also cannot differentiate between native and inactive forms. Thus localization using immunohistochemistry need not indicate the presence of activity in that compartment.
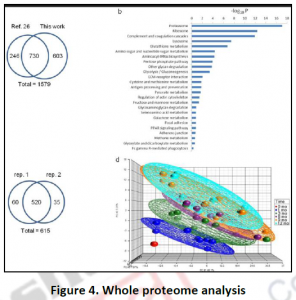
3.3. Proteome analysis by mass spectroscopy: Fractionation and immune-staining methods are low through put approaches with many limitations for investigating metabolic compartmentation. In the last few decades, targeted mass spectrometry combined with classical cell fractionation techniques helped the high through put cataloguing of proteomes of various organisms, cell compartments as well as various physiological conditions (Figure 4). Such whole proteome characterizations have not only confirmed the localizations of various proteins identified by previous studies but also revealed novel functions to already known or novel proteins.
Eg: In mitochondria, 20% of the identified proteins with unknown functions. Functional clustering of many novel proteins into various protein classes like kinases or phosphatases can also done. The reliability of the proteome result through any technique absolutely depends on the purity of the original organelle preparation and the level of cross contamination by proteins from other compartments. Similar to immune-histochemical technique, mass spectroscopic methods cannot distinguish between active and inactive proteins. It is challenging to identify hydrophobic proteins due to the difficulty in preparing highly purified membrane preparations like plasmalemma, ER, golgi membrane proteins etc.These existing limitations of spectroscopy can be tackled using isotope tagging and using laser microdissection capture microscopy.
3.4. Fluorescent protein tagging and other Invivo imaging.: The existing error pronicity due to microscopy, fractionation and spectroscopy based assays were tackled using fluorescent tagged affinity based molecules.This can be visualized by state of art fluorescent or confocal microscopy of processed samples as well as invivo live cell imaging (Figure 5). Using native promoter and other regulatory elements based expression of the target protein, exact location and distribution among cells or tissues can be analyzed. Fusion protein constructs using GFP(Green fluorescent protein),YFP (yellow fluorescent protein) etc has been initially used for pilot studies using transient and stable expression systems. Signal sequences or leader peptides were used for accurate targeting of these constructs to accurate locations like plastids, mitochondria etc. It’s also crucial to decide where to attach the fluorescent tag to the target protein- ‘C’ or the ‘N’ terminal. Due to the presence of signal peptide sequences for targeting to plastids, mitochondria etc on the ‘N’ terminal,tagging should be avoided at this end. ’C’ terminal also contain important targeting information such as endoplasmic reticulum retention signals. Thus by minimizing the disruption of targeting signals at both terminus, the default strategy is to insert fluorescent tags about 10 amino acids upstream to ‘C’ terminus, flanked by 6-9 amino acids Gly or Ala rich peptides for maximum integrity. This approach was successfully used for peroxisomal, tonoplast, plasma membrane, plasmodesmatal, cellwall, cytoskeletal, nuclear, proplastidial, cytosolic proteins etc.
NMR (Nuclear Magnetic resonance) spectroscopy is recently used to determine the intracellular distribution of inorganic orthophosphate and can detect changes in energy or PH of living cells. NMR has very much positively used for measuring flux rate of sucrose or water through phloem and xylem. In vivo NMR is poor in sensitivity and can only detect metabolites that are present in millimolar concentrations. A more accurate imaging approach is PET(Positron emission tomography) which detects short lived isotopes to label sugars and polyphenols. Nevertheless its very expensive and hence found very little applications.
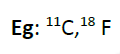
3.5. Computational or In silico approaches: A holistic approach of various proteomic platforms from fractionation to PET or fluorescent protein-labelling studies have revealed the target location for over thousands of protein in various plants. For example, 4000 proteins in Arabidopsis thaliana; and more than 24000 protein with unknown locations. To tackle this huge problem, and for annotating functions to unknown proteins certain In silico computational approaches have been developed based on primary protein sequence. It includes – TargetP, http://www.cbs.dtu . dk/services/TargetP/; Predotar, http://www.inra.fr/predotar/ ; iPSORT, http://hc.ims.u-tokyo.ac.jp/iPSORT/ ; SubLoc, http://www.bioinfo.tsinghua.edu.cn/SubLoc/. Most programs aims to identify the ‘C’ or ‘N’ terminus or signal peptide sequence for accurate location analysis. To access the specificity and sensitivity of prediction programs, several experimentally or predicted data sets are used. But most favorable assessments was found with at least 15% misidentified in case of plastid proteins.This misinterpretation can be improved with few false positives using several in silico prediction programs.
4. Plant metabolic pathways manifested through enzyme compartmentation
Over the last four decades, intense pain staking research on various platforms and techniques were carried out to understand the role of compartmentation in plant metabolism. Most of the primary metabolic pathways and their locations have been well explored and mapped along with few novel metabolic pathways.This proteomics paved a way towards localisomics which ended with interactomics. Lets discuss few pathways:-
4.1. Glycolysis: Central role of plastids in carbohydrate metabolism was well explored from 1970s to 1980s along with fatty acids, nucleotides and other secondary metabolism. Other studies revealed that separate isoforms of key enzymes in the pathway of sulphate assimilation and cysteine biosynthesis are found in chloroplasts, cytosol and mitochondria. A striking discovery of the existence of glycolysis and oxidative pentose phosphate pathway in duplicate manner both in cytosol and plastid stroma revealed how energy and carbon skeletons used up in both compartments. Another recent finding was the localization of most glycolytic enzymes in the cytosol, closely associated to outer mitochondrial membrane. The direct channeling of pyruvate into mitochondria for krebs cycle has also elucidated the need of micro-compartmentation. During photosynthesis in chloroplasts, several calvin cycle enzymes appear to form multi-enzyme complexes associated with membranes,where ATP and NADPH are synthesized.
4.2. Gluconeogenesis: Conversion of lipids to sugars in germinating oil seeds is another highly compartmentalized metabolic pathway-commonly known as the ‘glyoxylate cycle’.This is a tripartite pathway involving peroxisome,cytosol and mitochondria. Extensive studies done in castor bean seedings solved the existing questions like-how carbon is transported between these three organelles? What is the vital role of malate dehydrogenase?
It was interesting to observe that high NADH:NAD+ ratio build up in the peroxisome arising due to β-oxidation,would be unfavourable for the catalysis of malate dehydrogenase(MDH) reaction to proceed in the oxidative direction. Knock out mutants lacking MDH showed that the reaction proceeds in other direction also in line with the conclusion and clarified that oxaloacetate converts to malate and reoxidize the NADH produced by β-oxidation of fatty acids.
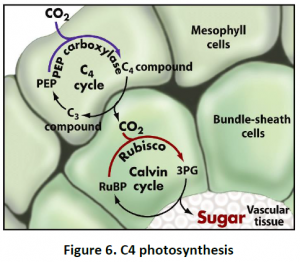
4.3. C4 Photosynthesis: Photosynthesis of C4 plants can be prevented by maintaining a high CO2 concentration at the site of fixation by Rubisco in bundle sheath cells. This pathway is unique in terms of compartmentation between two types of cells rather than only organelles.The regulation is tightly coordinated between mesophyll and bundlesheath cells as well as among chloroplast, cytosol and mitochondria. CO2 fixed initially in mesophyll cell cytoplasm by PEP carboxylase forms oxaloacetic acid, which further converts to malate or aspartate and is mobilized via plasmodesmata into bundle sheath cells. Malate or aspirate then decarboxylated in the chloroplasts or cytosol and mitochondria, releasing CO2 to be refixed by Rubisco. Other pathways are also compartmented between mesophyll and bundle sheath cells in C4 plants. For example, Sucrose is synthesized only in mesophyll cells, while starch synthesis is restricted to bundle sheath cells (Figure 6).
Conclusion:
Compartmentation is a way of localization and regulation. Various methods of compartmentation includes cell fractionation, immunohistochemistry, proteome analysis by mass spectroscopy, fluorescent protein tagging and other in vivo imaging and computational or in silico approaches. Most of the primary metabolic pathways and their locations have been well explored and mapped along with few novel metabolic pathways like glycolysis, gluconeogenesis and C4 photosynthesis. This proteomics paved a way towards localisomics which ended with interactomics explains the complexity of plant metabolism.
| you can view video on Enzyme compartmentalization in cells and tissues |
